ML estimation of the Matrix Variate t Distribution
Geoffrey Thompson
2024-09-30
Source:vignettes/matrix-t-estimation.Rmd
matrix-t-estimation.RmdEstimation of the Matrix Variate t Distribution
The parameters of the multivariate t distribution can be estimated using the EM algorithm. An EM algorithm for the multivariate -distribution with no missing data was provided by Rubin (1983). This was variously extended and refined, as the EM algorithm can be quite slow to converge. One set of refinements split the M-step into a series of conditional maximization (CM) steps (Meng and Rubin 1993), and then a later refinement (called ECME - Expectation/Conditional Maximization Either) allowed for either conditional maximization steps or maximizing the constrained actual likelihood (Liu and Rubin 1994), which leads to dramatically faster convergence. The matrix variate is an extension of the multivariate version (Dickey 1967); however, unlike the normal distribution, it cannot be treated as a rearrangement of the multivariate case, so the addition of the extra dimension poses a non-trivial problem for extending the results.
An random matrix is distributed as a matrix variate random variable if it has probability density function as follows: $$f({\mathbf {X}};\nu,{\mathbf {M}}, {\boldsymbol \Omega }, {\boldsymbol \Sigma }) = {\frac {\Gamma _{p}\left({\frac {\nu +n+p-1}{2}}\right)}{(\pi )^{{\frac {np}{2}}}\Gamma _{p}\left({\frac {\nu +p-1}{2}}\right)}}|{\boldsymbol \Omega }|^{{-{\frac {n}{2}}}}|{\boldsymbol \Sigma }|^{{-{\frac {p}{2}}}}\times \left|{\mathbf {I}}_{n}+{\boldsymbol \Sigma }^{{-1}}({\mathbf {X}}-{\mathbf {M}}){\boldsymbol \Omega }^{{-1}}({\mathbf {X}}-{\mathbf {M}})^{{{\rm {T}}}}\right|^{{-{\frac {\nu +n+p-1}{2}}}}$$
With
and
covariance matrices of appropriate dimension,
a mean matrix,
the degrees of freedom parameter, and
the multivariate gamma function, which is implemented in the CholWishart
package available on CRAN.
For and (or and ), this reduces to the familiar multivariate distribution. If the dimensional random variable follows the matrix variate distribution with center , positive definite spread matrices () and (), and degrees of freedom , it can be shown that, given a certain weight matrix , has a matrix variate normal distribution and that is Wishart-distributed. Specifically, Additionally, it can be shown the Wishart is the conjugate prior distribution for the parameter , and thus the conditional posterior distribution of , i.e., its distribution given is
For an observed set of , , then, we can construct an ECM-style algorithm by augmenting our data with a set of weights, , and use this to estimate the parameters of the matrix variate distribution. Briefly, we define a set of complete data sufficient statistics based on the and update them with their expectations in the E-step. Then, the other parameters are maximized in the CM steps.
The complete data sufficient statistics for the quantities to be estimated are:
E-step
Define at step the set . Then, given these values, we have, based on the properties of the Wishart distribution, From this we can derive the other expected values: This last value only needs to be computed if is unknown:
CM Steps
Similarly to the case of the multivariate , it is more efficient to partition the maximization step into multiple steps. This is an ECME algorithm that first maximizes the expected log-likelihood for and then maximizes the actual log-likelihood over given , similar to .
Based on the updated weight matrices and statistics based on and ,
And, again, with the set of observed, the MLE of can be obtained: Note that . Specifically, we have: where is the appropriate statistic with factored out and is the -dimensional digamma function. This can be solved for using a 1-dimensional search.
Applications and results
With known
If the degrees of freedom parameter,
known, the estimation is fairly straightforward. The procedure is
similar to the multivariate
or the matrix variate normal. In this case, the interface is just like
the interface for the MLmatrixnorm() function:
set.seed(20190622)
sigma = (1/7) * rWishart(1, 7, 1*diag(3:1))[,,1]
A = rmatrixt(n=100,mean=matrix(c(100,0,-100,0,25,-1000),nrow=2),
V = sigma, df = 7)
results=MLmatrixt(A, df = 7)
print(results)
#> $mean
#> [,1] [,2] [,3]
#> [1,] 99.8291911 -1.000708e+02 25.01042
#> [2,] -0.0425028 -8.261356e-03 -1000.02357
#>
#> $U
#> [,1] [,2]
#> [1,] 1.00000000 -0.08039981
#> [2,] -0.08039981 0.97069319
#>
#> $V
#> [,1] [,2] [,3]
#> [1,] 1.0000000 0.21375613 0.11094767
#> [2,] 0.2137561 0.14872070 0.04098332
#> [3,] 0.1109477 0.04098332 0.12860660
#>
#> $var
#> [1] 5.148569
#>
#> $nu
#> [1] 7
#>
#> $iter
#> [1] 44
#>
#> $tol
#> [1] 1.155394e-08
#>
#> $logLik
#> [1] -386.7623
#>
#> $convergence
#> [1] TRUE
#>
#> $call
#> MLmatrixt(data = A, df = 7)There are two restrictions possible for the mean matrices:
row.mean = TRUE will force a common mean within a row and
col.mean = TRUE will force a common mean within a column.
Setting both will ensure a constant mean for the entire system.
Restrictions on
and
,
the row-wise variance and column-wise variance, are possible with
row.variance and col.variance commands.
The options for variance restrictions are the same as for the
MLmatrixnorm() function. Currently the options for variance
restrictions are to impose an AR(1) structure by providing the
AR(1) option, a compound symmetry structure by providing
the CS option, to impose a correlation matrix structure by
specifying correlation or corr, or to impose
an identical and independent structure by specifying
Independent or I. This works by using
uniroot to find the appropriate
which sets the derivative of the log-likelihood to zero for the
AR and CS options - it is not fast but if this
is the true structure it will be better in some sense than an
unstructured variance matrix. The
parameter should be
and is forced to be non-negative. If the data behaves incompatibly with
those restrictions, the function will provide a warning and exit with
the current model fit.
With unknown
Estimation of
,
the degrees of freedom parameter, is slow and the principal mathematical
difficulty of the matrix-variate
distribution. It is performed using ECME. Generally, a fair amount more
data are needed in order to have good convergence properties for the
estimator, but they have not been derived analytically. Here you can see
the recovery of the parameter for a few sample sizes. Because of the
relative slowness of running a longer simulation, this only includes one
set of examples. I give code for a longer and larger simulation than
what is plotted if you’re really interested below. What is plotted below
is only the df = 10 example with 75 trials and a maximum
number of iterations max.iter = 20. The full simulation
may take several minutes.
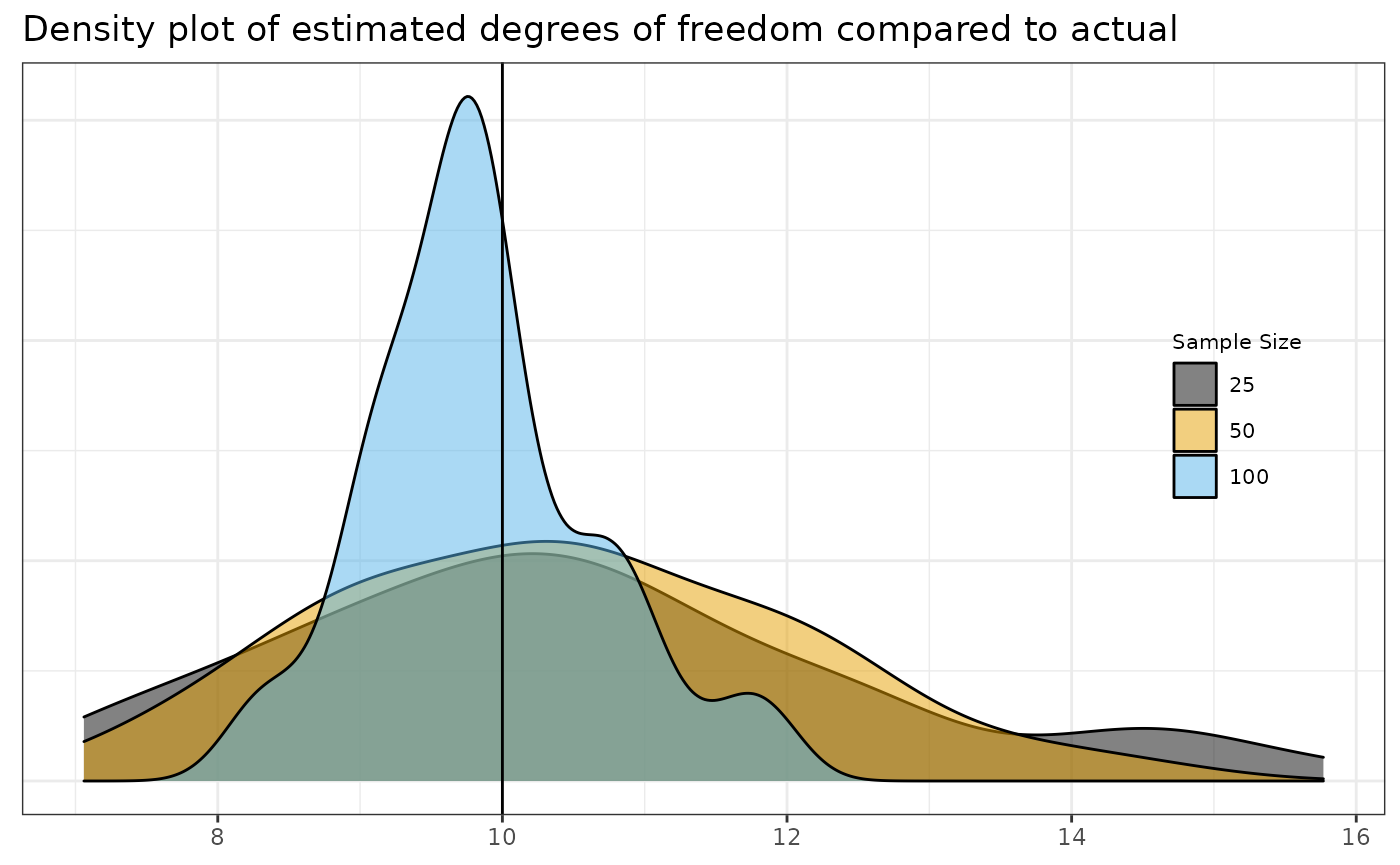
| samplesize | min | max | median | mean | sd |
|---|---|---|---|---|---|
| 25 | 7.059192 | 15.76929 | 10.316514 | 10.512701 | 2.0603677 |
| 50 | 7.220806 | 14.47148 | 10.424858 | 10.415426 | 1.6347640 |
| 100 | 8.281614 | 11.85454 | 9.826301 | 9.859941 | 0.8019631 |
As expected, increased sample size leads to better results in recovering the parameter. The results for smaller sample sizes would be more divergent if left to run until convergence.
Use for classification
Using the
distribution works in both matrixlda() and
matrixqda() as expected by specifying
method = "t" and providing either a single parameter (for
lda or qda) for the degrees of freedom or a
vector as long as the number of classes (for qda).
Additional parameters for fitting can be passed through the
... to MLmatrixt() just as for the normal
case, including estimating the degrees of freedom parameter. The
qda will only estimate nu with it varying
between groups, it will not estimate a common nu.
A <- rmatrixt(30, mean = matrix(0, nrow=2, ncol=3), df = 10)
B <- rmatrixt(30, mean = matrix(c(1,0), nrow=2, ncol=3), df = 10)
C <- rmatrixt(30, mean = matrix(c(0,1), nrow=2, ncol=3), df = 10)
ABC <- array(c(A,B,C), dim = c(2,3,90))
groups <- factor(c(rep("A",30),rep("B",30),rep("C",30)))
prior = c(30,30,30)/90
matlda <- matrixlda(x = ABC,grouping = groups, prior = prior,
method = 't', nu = 10, fixed = TRUE)
predict(matlda, newdata = ABC[,,c(1,31,61)])
#> $class
#> [1] A B C
#> Levels: A B C
#>
#> $posterior
#> [,1] [,2] [,3]
#> [1,] 9.997064e-01 2.849389e-04 8.693417e-06
#> [2,] 4.174839e-05 9.999508e-01 7.439619e-06
#> [3,] 8.908508e-06 4.857627e-06 9.999862e-01Session info
This vignette was built using rmarkdown.
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] magrittr_2.0.3 dplyr_1.1.4 ggplot2_3.5.1 MixMatrix_0.2.8
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.5 jsonlite_1.8.9 highr_0.11
#> [4] compiler_4.4.1 tidyselect_1.2.1 Rcpp_1.0.13
#> [7] jquerylib_0.1.4 systemfonts_1.1.0 scales_1.3.0
#> [10] textshaping_0.4.0 yaml_2.3.10 fastmap_1.2.0
#> [13] R6_2.5.1 labeling_0.4.3 generics_0.1.3
#> [16] knitr_1.48 tibble_3.2.1 desc_1.4.3
#> [19] munsell_0.5.1 bslib_0.8.0 pillar_1.9.0
#> [22] rlang_1.1.4 utf8_1.2.4 cachem_1.1.0
#> [25] xfun_0.47 fs_1.6.4 sass_0.4.9
#> [28] cli_3.6.3 withr_3.0.1 pkgdown_2.1.1
#> [31] digest_0.6.37 grid_4.4.1 CholWishart_1.1.2.1
#> [34] lifecycle_1.0.4 vctrs_0.6.5 evaluate_1.0.0
#> [37] glue_1.7.0 farver_2.1.2 ragg_1.3.3
#> [40] fansi_1.0.6 colorspace_2.1-1 rmarkdown_2.28
#> [43] tools_4.4.1 pkgconfig_2.0.3 htmltools_0.5.8.1All the code for easy copying
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>"
)
set.seed(20190622)
sigma = (1/7) * rWishart(1, 7, 1*diag(3:1))[,,1]
A = rmatrixt(n=100,mean=matrix(c(100,0,-100,0,25,-1000),nrow=2),
V = sigma, df = 7)
results=MLmatrixt(A, df = 7)
print(results)
### Here is the long simulation
library(ggplot2)
set.seed(20181102)
df = c(5, 10, 20)
df5 <- rep(0,200)
df10 <- rep(0,200)
df100 <- rep(0,200)
df550 <- rep(0,200)
df1050 <- rep(0,200)
df2050 <- rep(0,200)
df5100 <- rep(0,200)
df10100 <- rep(0,200)
df20100 <- rep(0,200)
meanmat = matrix(0,5,3)
U = diag(5)
V = diag(3)
for(i in 1:200){
df5[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 5, n = 35, U =U, V =V), fixed = FALSE)$nu
df10[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 10, n = 35, U =U, V =V), fixed = FALSE)$nu
df100[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 20, n = 35, U =U, V =V), fixed = FALSE)$nu
df550[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 5, n = 50, U =U, V =V), fixed = FALSE)$nu
df1050[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 10, n = 50, U =U, V =V), fixed = FALSE)$nu
df2050[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 20, n = 50, U =U, V =V), fixed = FALSE)$nu
df5100[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 5, n = 100, U =U, V =V), fixed = FALSE)$nu
df10100[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 10, n = 100, U =U, V =V), fixed = FALSE)$nu
df20100[i] = MLmatrixt(rmatrixt(mean = meanmat,
df = 20, n = 100, U =U, V =V), fixed = FALSE)$nu
}
truedataframe = data.frame(truedf = factor(c(5,10,20),
label = c('5 df', '10 df', '20 df')),
estdf = c(5,10,20))
dfdataframe = data.frame(truedf = factor(rep(rep(c(5,10,20), each = 200),3),
label = c('5 df', '10 df', '20 df')),
estdf = c(df5, df10, df100, df550, df1050, df2050, df5100, df10100, df20100),
samplesize = factor(rep(c(35,50,100), each = 600)))
library(tidyverse)
denseplot <- ggplot(data = subset(dfdataframe, estdf < 200),
aes(x=estdf, fill=samplesize)) +
geom_density(alpha = .5) +
geom_vline(data = truedataframe,
mapping = aes(xintercept = estdf),
size = .5) +
theme_bw() +
theme(axis.ticks.y=element_blank(), axis.text.y=element_blank(),
strip.text = element_text(size = 8),
legend.justification=c(1,0), legend.position=c(.95,.4),
legend.background = element_blank(),
legend.text =element_text(size = 8), legend.title = element_text(size = 8)) +
ggtitle("Density plot of estimated degrees of freedom compared to actual") +
xlab(NULL) +
ylab(NULL) +
scale_fill_manual(values = c("#050505", "#E69F00", "#56B4E9"),
name = "Sample Size") +
facet_wrap(factor(truedf)~., scales="free") +
NULL
denseplot
knitr::kable(dfdataframe %>% group_by(truedf, samplesize) %>%
summarize(min = min(estdf), max = max(estdf),
median = median(estdf),
mean=mean(estdf),
sd = sd(estdf)))
### Here ends the long simulation
##### Here is what is really run
set.seed(20190621)
df10 <- rep(0,50)
df1050 <- rep(0,50)
df10100 <- rep(0,50)
for(i in 1:50){
df10[i] = suppressWarnings(MLmatrixt(rmatrixt(mean = matrix(0,5,3),df = 10, n = 25), fixed = FALSE, df = 5, max.iter = 20)$nu)
df1050[i] = suppressWarnings(MLmatrixt(rmatrixt(mean = matrix(0,5,3),df = 10, n = 50), fixed = FALSE, df = 5, max.iter = 20)$nu)
df10100[i] = suppressWarnings(MLmatrixt(rmatrixt(mean = matrix(0,5,3),df = 10, n = 100), fixed = FALSE, df = 5, max.iter = 20)$nu)
}
dfdataframe = data.frame(label = c('10 df'),
estdf = c(df10, df1050, df10100),
samplesize = factor(rep(c(25,50,100), each = 50)))
library(ggplot2)
library(dplyr)
library(magrittr)
denseplot <- ggplot(data = subset(dfdataframe, estdf < 200),aes(x=estdf, fill=samplesize)) +
geom_density(alpha = .5) +
geom_vline(mapping = aes(xintercept = 10),
size = .5) +
theme_bw() +
theme(axis.ticks.y=element_blank(), axis.text.y=element_blank(), strip.text = element_text(size = 8),
legend.justification=c(1,0), legend.position=c(.95,.4),
legend.background = element_blank(),
legend.text =element_text(size = 8), legend.title = element_text(size = 8)) +
ggtitle("Density plot of estimated degrees of freedom compared to actual") +
xlab(NULL) +
ylab(NULL) +
scale_fill_manual(values = c("#050505", "#E69F00", "#56B4E9"), name = "Sample Size") +
# facet_wrap(factor(truedf)~., scales="free") +
NULL
denseplot
knitr::kable(dfdataframe %>% group_by(samplesize) %>%
summarize(min = min(estdf), max = max(estdf),
median = median(estdf),
mean=mean(estdf),
sd = sd(estdf)))
#### Here ends what is really run
A <- rmatrixt(30, mean = matrix(0, nrow=2, ncol=3), df = 10)
B <- rmatrixt(30, mean = matrix(c(1,0), nrow=2, ncol=3), df = 10)
C <- rmatrixt(30, mean = matrix(c(0,1), nrow=2, ncol=3), df = 10)
ABC <- array(c(A,B,C), dim = c(2,3,90))
groups <- factor(c(rep("A",30),rep("B",30),rep("C",30)))
prior = c(30,30,30)/90
matlda <- matrixlda(x = ABC,grouping = groups, prior = prior,
method = 't', nu = 10, fixed = TRUE)
predict(matlda, newdata = ABC[,,c(1,31,61)])
sessionInfo()